ACS-SWRM-2023: Dynamic Perspectives on Catalytic Function in Adenylate Kinase
DISCLAIMER: The content in this article is for demonstration purposes only and may contain errors and technical inaccuracies.
This article aims to provide a concise overview of my presentation at the ACS-SWRM-2023 conference, organized by the American Chemical Society SWRM Board in November 2023, held in Oklahoma City, OK, USA.
The presentation was based on the study which is an extension of the following studies from Nam group:
Nam, K.; Arattu Thodika, A. R.; Grundström, C.; Sauer, U. H.; Wolf-Watz, M. Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release. J. Chem. Inf. Model. 2023.
Dulko-Smith, B.; Ojeda-May, P.; Ådén, J.; Wolf-Watz, M.; Nam, K. Mechanistic Basis for a Connection between the Catalytic Step and Slow Opening Dynamics of Adenylate Kinase. J. Chem. Inf. Model. 2023, 63 (5), 1556–1569.
Ojeda-May, P.; Mushtaq, A. U. I.; Rogne, P.; Verma, A.; Ovchinnikov, V.; Grundström, C.; Dulko-Smith, B.; Sauer, U. H.; Wolf-Watz, M.; Nam, K. Dynamic Connection between Enzymatic Catalysis and Collective Protein Motions. Biochemistry 2021, 60 (28), 2246–2258.
Catalytic Mechanism of Adenylate Kinase
Adenylate kinase (Adk) is a ubiquitous enzyme that catalyses the interconversion between ATP+AMP+Mg2+ and 2ADP+Mg2+. The catalytic mechanisk which include conformational changes and ligand binding events is shown in the following figure:
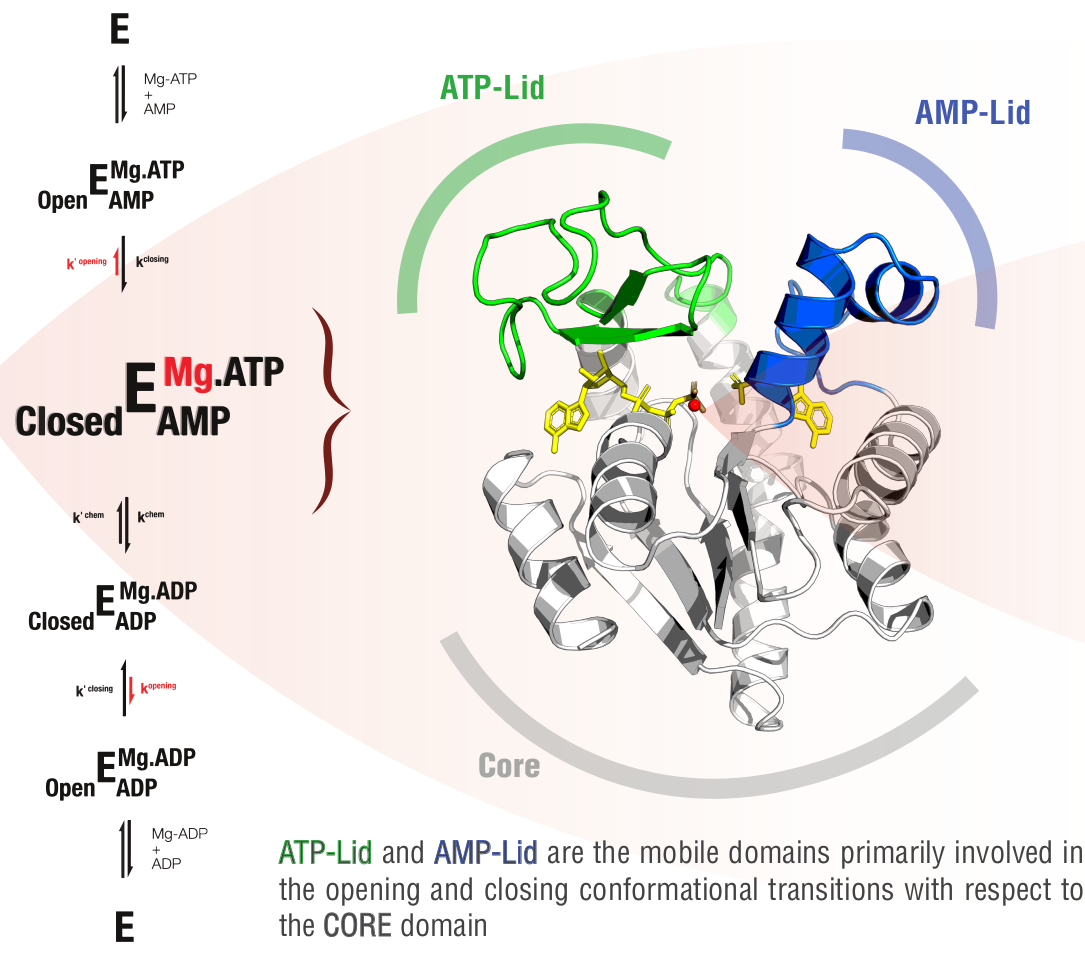 Schematic representation of Adk mechanism and different domains involved in conformational transitions
Schematic representation of Adk mechanism and different domains involved in conformational transitions
Dual Role of Active Site Residues in Adk
The active site surrounding the ligand, depicted as yellow sticks in the figure above, comprises highly charged arginine residues that exhibit a high degree of conservation across ancestral species. The referenced articles provide comprehensive insights into the dual role of these arginine residues, detailing their involvement in both close-to-open conformational transitions and catalytic processes. An extension to this study, encompassing further details, is slated for publication in the near future.
Gaussian Mixed Model (GMM) to Study Connection between Conformational Transitions and Catalysis
While numerous methods exist in the literature for clustering Molecular Dynamics (MD) trajectories to investigate protein conformational transitions, selecting an analysis method that yields meaningful insights in a reduced dimensionality poses a considerable challenge. A thorough understanding of adenylate kinase conformational transitions facilitates the effective application of Gaussian Mixture Model (GMM) to explore the interplay between conformational changes and catalysis.
The application of a Gaussian-based model necessitates near-to-full coverage of the protein’s conformational space, a task not always feasible with MD simulations alone. To address this, focusing on small time-scale motions and employing numerous replicas of MD simulations can provide a reasonable coverage of the conformational space within a limited timeframe. Alternatively, enhanced sampling methods can also be employed. In our study, we adopted the former approach to generate the conformational space of the protein.
Establishing a direct connection between conformational transitions and catalysis is inherently challenging. However, we employed a sequential approach, utilizing Gaussian-based Kernel Density Estimation (KDE) clustering on a selected feature influencing catalytic effects. Subsequently, we applied GMM on the clustered data based on a different feature influencing conformational transitions. This approach offers valuable insights into the connection between conformational transitions and catalysis.
An upcoming update to this article will provide detailed insights into the application of GMM for studying the connection between conformational transitions and catalysis utilizing Python libraries such as scikit-learn, PyMOL, and MDAnalysis.
